Part:BBa_K3249008
CRY2-VPR
This part is composed of Cryptochrome 2 (CRY2) photoreceptor linked to a VPR transcriptional activator intended for use in the light activated CRISPR/Cas9 effector system (LACE). When induced by blue light, CRY2 dimerizes with its binding partner CIBN, effectively bringing the transcriptional activator VPR to the target site defined by the dCas9/gRNA complex. gRNAs can be designed in order for the dCas9 protein to target and increase transcription of specific genes. A figure of how the system works is shown below.

|
| Figure 1: Light activation triggers the dimerization of CRY2 to CIBN, which brings the activator VPR to the promoter region of the GOI increasing rate of transcription. |
Characterization
We used lipofectamine to transiently cotransfect CHO cells with 2 plasmids. One containing CIBN-dCas9-CIBN and our 3 gRNAs and the other containing CRY2-VPR. When both plasmids are successfully transfected into the cells we expect to see an increase in transcription for the gene IL1RN when the cells are induced by blue light. The cells were incubated at 37° C for 24 hours to allow them to adhere to the 24 well plate. After 24 hours, they were placed into our light plate apparatus (LPA) while still being incubated at 37°C. The LPA illuminated the wells with blue light for 24 hours. We then extracted the RNA from the CHO cells and used reverse transcriptase to convert this RNA into cDNA. Using the cDNA as template DNA, qPCR was performed to quantify the fold increase of IL1RN expression compared to untreated cells. The data below compares the effectiveness of our CRY2-VP64 to NEU China 2016s CRY2-VP64 (Part:BBa_K1982010) and our activator CRY2-VPR.
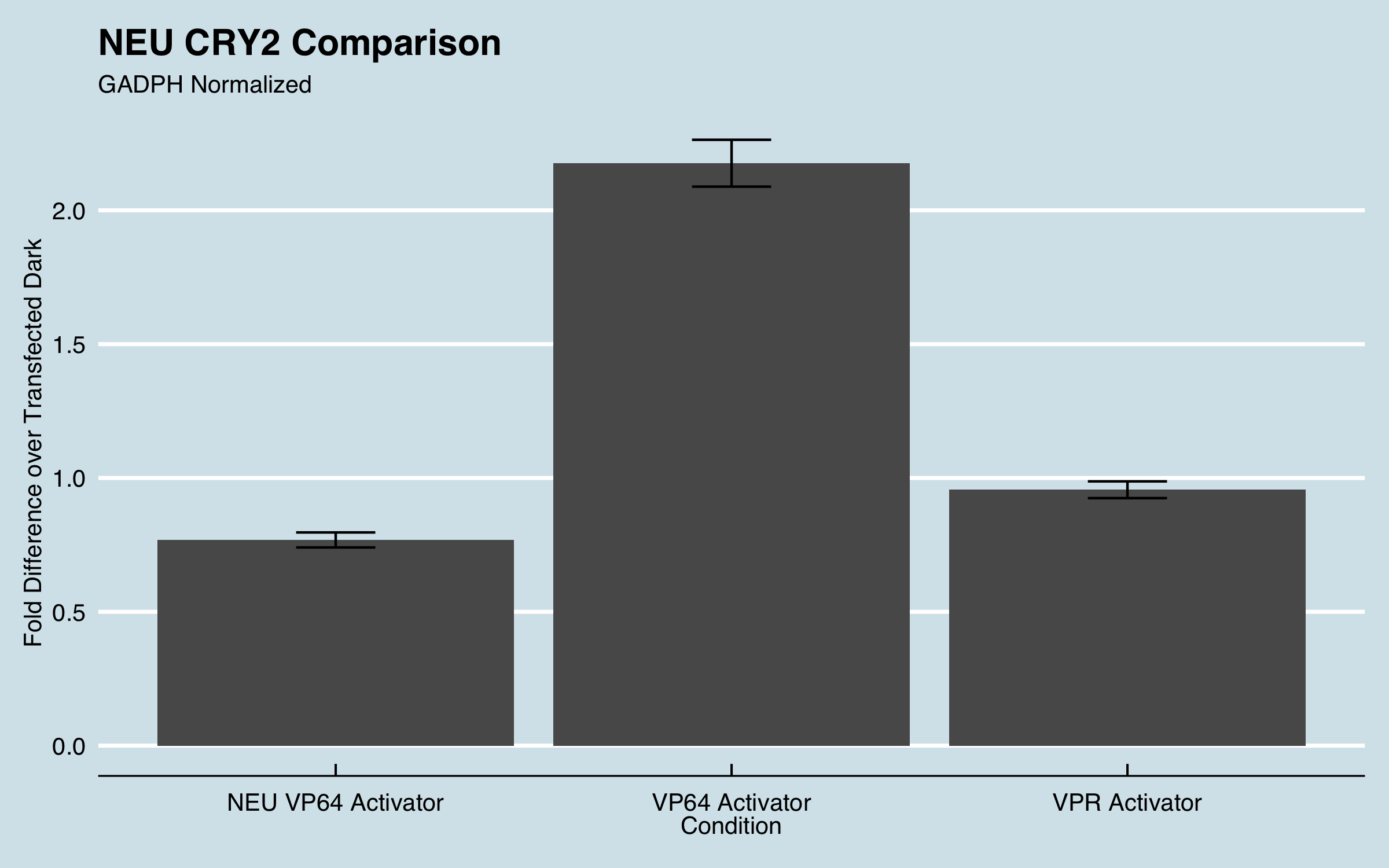
|
| Figure 1: Fold expression of IL1RN in CHO cells when transfected with different transcriptional activators. |
Fold increase was calculated using the delta-delta Ct method for qPCR. All data was normalized to the reference genes recommended by MIQE guidelines and fold increase was compared to untransfected cells. For more information about our experimental design and data analysis refer to our wiki (https://2019.igem.org/Team:UC_Davis).
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal EcoRI site found at 866
Illegal EcoRI site found at 3650
Illegal XbaI site found at 2226
Illegal PstI site found at 676
Illegal PstI site found at 2733
Illegal PstI site found at 2739 - 12INCOMPATIBLE WITH RFC[12]Illegal EcoRI site found at 866
Illegal EcoRI site found at 3650
Illegal NheI site found at 33
Illegal PstI site found at 676
Illegal PstI site found at 2733
Illegal PstI site found at 2739 - 21INCOMPATIBLE WITH RFC[21]Illegal EcoRI site found at 866
Illegal EcoRI site found at 3650
Illegal BglII site found at 536
Illegal BglII site found at 995
Illegal BglII site found at 2527
Illegal BamHI site found at 1474
Illegal BamHI site found at 1987
Illegal BamHI site found at 3296
Illegal BamHI site found at 3341 - 23INCOMPATIBLE WITH RFC[23]Illegal EcoRI site found at 866
Illegal EcoRI site found at 3650
Illegal XbaI site found at 2226
Illegal PstI site found at 676
Illegal PstI site found at 2733
Illegal PstI site found at 2739 - 25INCOMPATIBLE WITH RFC[25]Illegal EcoRI site found at 866
Illegal EcoRI site found at 3650
Illegal XbaI site found at 2226
Illegal PstI site found at 676
Illegal PstI site found at 2733
Illegal PstI site found at 2739
Illegal NgoMIV site found at 2703
Illegal NgoMIV site found at 3108
Illegal AgeI site found at 420
Illegal AgeI site found at 1149
Illegal AgeI site found at 3237 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 772
Illegal BsaI.rc site found at 20
Illegal BsaI.rc site found at 181
Illegal SapI.rc site found at 289
| None |
